Server Manual
1. General Information
Short Linear Motifs (SLiMs, motifs) are typically short sequence stretches of 3–10 amino acids that mediate protein–protein interactions. These segments are often described using regular expressions that capture sequence definitions but do not consider biological or biophysical properties; consequently, they yield many false positives.
The SLiMMine resource is a collection of predicted SLiMs in the human proteome. We developed a prediction method to identify motifs across the human proteome. SLiMMine integrates known motifs as defined in the Eukaryotic Linear Motif resource and also highlights short sequence segments for which prediction probabilities indicate functional motifs, even when no regular expression has been defined for that region.
In addition, we refined and extended the annotations of interaction partners and Gene Ontology terms for all motifs to enable better result filtering. For more information about the methodology, please refer to the descriptive article.
2. Navigation on the Webpage
2.1. Search Options
There are three ways to search for motifs in the SLiMMine resource.
2.1.1 Search for protein:
Proteins can be searched by gene name, UniProt accession (AC), or UniProt ID.
The search results page displays the following information for each hit: gene name, UniProt accession and identifier, and localization from UniProt. It also shows the total number of motifs, high-confidence motifs, experimental motifs, and experimental protein–protein interactions. More detailed information is available on the protein entry page, which is linked from the results page.
2.1.2. Search for motif class:
SLiMs can be searched by their ELM class name.
2.1.3. Prediction:
Custom regular expressions can be searched. When searching using regular expressions, the environment of the motif (i.e., intracellular or extracellular) must be specified.
When searching for a motif class or a custom regular expression, the results page displays a table containing the following information: gene name, UniProt AC, start position, end position, sequence, prediction probability, and localization. More information is available on the linked protein entry page.
2.2. Protein Entry Page
The protein entry page aggregates information about SLiMs and protein interaction partners mediated by these SLiMs.
2.2.1. General
The General section displays the following information: gene name, UniProt accession, and UniProt identifier. Localization information (from UniProt) and PFAM domains are also shown. Summary statistics, including the total number of motifs, high-confidence motifs, experimental motifs, and experimental protein–protein interactions, are provided.
2.2.2. Motifs
The Motifs section relies heavily on the SLiMMine prediction algorithm and on ELM motif classes. The localization(s), binding domain(s), possible interaction partners, and Gene Ontology terms of the motif classes have been manually refined to enable the more precise filtering of the corresponding motif hits and the potential interactions mediated by them.
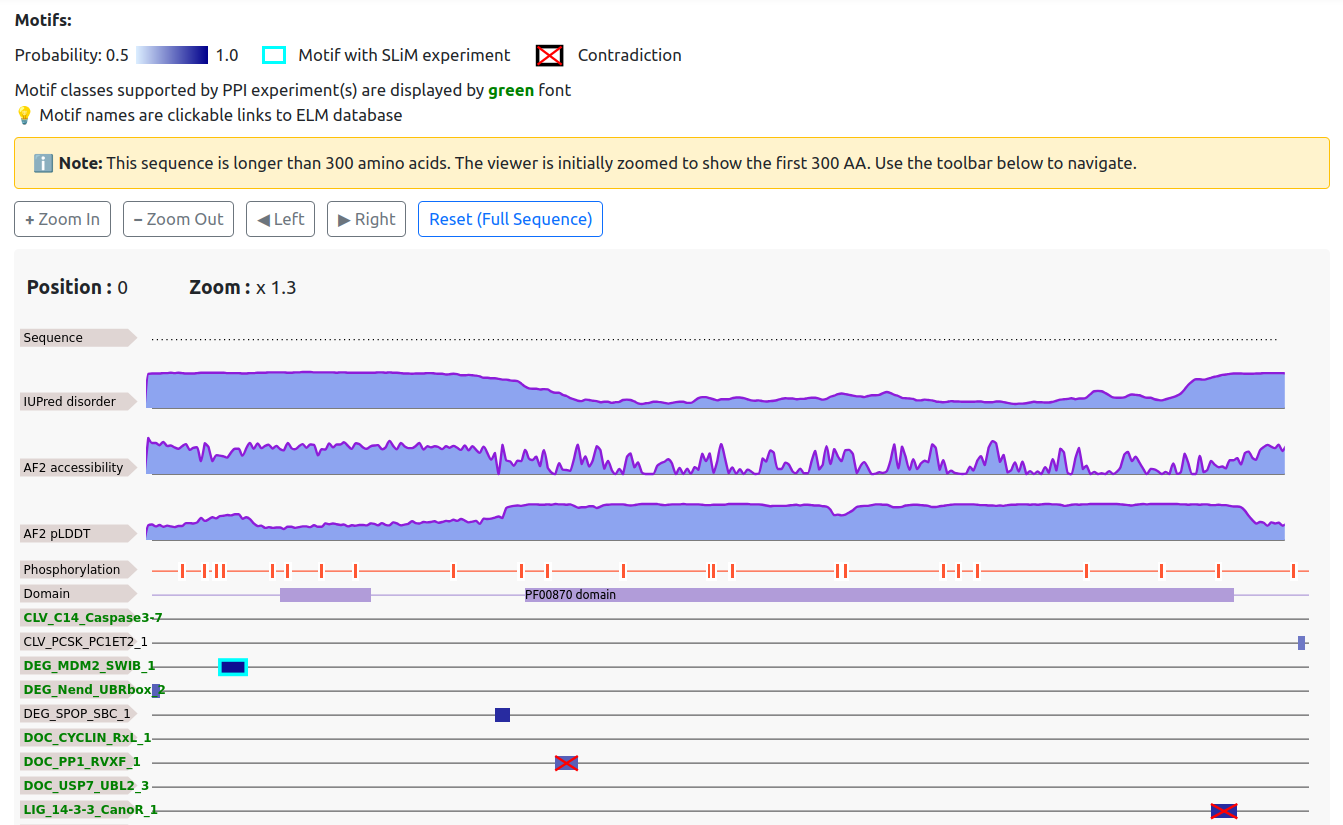
This section summarizes all potential motif-mediated interactions and short linear motifs. Motifs tend to favor accessible and disordered regions and are less likely to occur within globular domains. Post-translational modifications can themselves be considered motifs; in particular, phosphorylation often acts as a regulatory switch that must be present alongside another SLiM for functionality.
Accordingly, the chart not only highlights motifs but also displays AlphaFold2 confidence scores, accessibility values associated with protein disorder, AIUPred disorder predictions, experimental phosphorylation sites from PhosphoSitePlus and a large-scale study by Ochoa et al., PFAM domains, and transmembrane topology .
Below this general information, SLiMs from ELM are listed. Motifs defined by more stringent criteria (i.e., with lower pattern probability) are shown separately at the top. If the entry protein has experimentally verified interactions with known binding partners of a motif, this is highlighted in green in the list on the left.
The intensity of the blue boxes representing motifs corresponds to their prediction probability. Motifs with probabilities below 0.5 are displayed only if experimental evidence exists. A cyan frame indicates experimental evidence supporting a motif, while red crosses indicate unfavorable annotations. When searching for regular expressions or motif classes, the query is always displayed at the top of the sequence chart.
On the left side of the chart, motif classes and transmembrane topology entries link to the corresponding UniTmp or ELM pages for additional information. Clicking on a motif jumps to the corresponding entry in the motif details table.
2.2.3. Motif Details
The motif details table provides detailed information corresponding to the chart above:
- Class: ELM classification. Motifs with high pattern probability (i.e., less specific regular expressions that may yield many false positives) are marked with an asterisk (*).
- Motif sequence
- Start and end positions
- Prediction probability (SLiMMine prediction)
- Protein disorder AIUPred
- Conservation: Calculated using Chordata alignments after searching the regular expression within ±50 residues relative to the human position.
- Experimental evidence: From ELM, hLMPID, and PDB via 3did.
- Binding domains: Using InterPro.
- Notes: Highlight favorable and contradictory information, including:
- Whether the motif lies within a domain or a transmembrane region (shown in red). Domains were retrieved by InterProScan, considering PFAM entries that contain 'domain', and do not contain "motif" in their name. GO terms are categorized as necessary or favorable. Missing necessary terms are shown in red; missing favorable terms are shown in yellow; matching terms are shown in green.
- Whether the protein has experimentally verified interactions with listed SLiM binding partners (shown in green as “experimental PPI with partner(s)”).
- Whether the SLiM is functional only in transmembrane proteins. If the protein contains a transmembrane region, this is shown in green (“transmembrane protein”); otherwise, in red (“non-transmembrane protein”).
- Whether the motif lies within a domain or a transmembrane region (shown in red).
- Whether the motif is located in an ordered region (shown in red).
- Broad protein localization: if the motif is functional in cytosolic or extracystolic environments and the protein region matches this localization, it is shown in green; otherwise, in red.
- Some motifs are restricted to proteins containing a specific domain; absence of this domain is shown in red (“missing: domain”).
- Less defined motifs: these motifs have high pattern probability, which means they occur much more frequently in the proteome.
The table can be filtered by columns or restricted to experimental motifs or high-confidence motifs (prediction probability ≥ 0.9). Motifs are sorted based on supporting evidence.
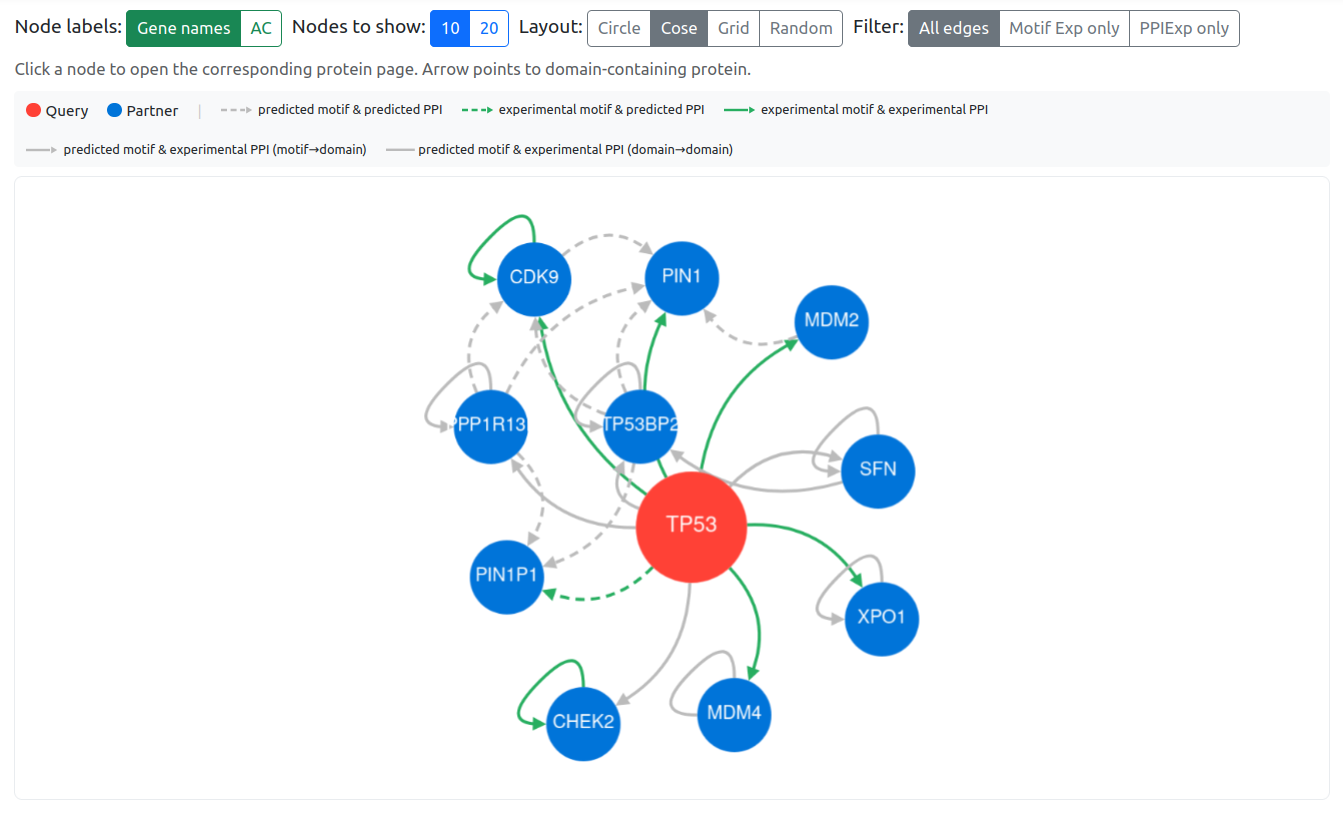
2.2.4. Protein Interaction Network
The protein interaction network is based on the refined partner lists for all SLiMs. Rather than relying solely on PFAM and InterPro domain definitions, curated lists of partner proteins were generated to reduce nonspecific domain-based associations.
The network displays the top 10 or 20 SLiM-mediated interactions for the entry protein. Arrows point toward the domain-containing binding partner. Grey dashed arrows indicate predicted interactions (both the motif within the protein and interaction wit the partner are predicted). Green dashed arrows indicate experimental evidence for the motif but not for the interaction partner. Green dashed arrows indicate predicted motif with experimental evidence on the protein interaction partner. Green solid arrows indicate experimental evidence for both the motif and the interaction partner (from ELM, LMPID, BIOGRID, PDB, and IntAct). Grey solid lines represent additional experimental protein–protein interactions within the network (based on BIOGRID, PDB, and IntAct).
2.2.5. Protein Interaction Details
The protein interaction details table summarizes potential motif-mediated interactions and their sources:
- Motif class mediating the interaction
- Gene name of potential partner
- Motif-level experimental (if any): ELM, LMPID, and PDB via 3did)
- Protein–protein interaction evidence (if any): ELM, LMPID , BIOGRID, PDB and IntAct
Potential interactions are sorted based on supporting evidence.
3. Downloads
3.1. Protein Entries
Entries can be downloaded from the protein entry page.
- Motif prediction results: motif class, start and end positions, prediction probability
- Interactions: motif class and binding partner
3.2. Motif Classes
Predictions for a motif class can be downloaded from the search page (UniProt AC, start, end, prediction probability).
3.3. Regular Expression Search Results
Predictions for regular expression searches can be downloaded from the search page (UniProt AC, start, end, prediction probability).